Background: Plasmacytoid dendritic cells (pDCs), being a primary source of natural type I interferon (IFN-I), have pivotal roles in orchestrating the immune response. Typically, only less than 1% pDCs can be detected in the total bone marrow or peripheral blood nuclear cells. However, the uncontrollable proliferation of transformed pDCs is a remarkable characteristic of blastic plasmacytoid dendritic cell neoplasm (BPDCN). Recently, an abnormal amplification of pDCs in myeloid neoplasms has also been reported in MDS/AML patients (pDC-AML/MDS). This abnormal increase in pDCs could potentially promote disease progression. In this study, we utilized single-cell RNA-sequencing (scRNA-seq) to examine the characteristics of pDCs in pDC-AML.
Method: Our study employed a mixed retrospective and prospective design. 85 patients diagnosed with PDC-AML/MDS/BPDCN, including 1 pDC-MDS, 17 pDC-AML, 50 non-pDC-AML (AML patients without pDCs expansion), 17 BPDCN from Tongji Hospital of Huazhong University of Science and Technology in Wuhan, China, between July 2014 and June 2023. scRNA-seq using 10x Genomics platform were performed on bone marrow (BM) samples from 8 pDC-AML patients and 1 BPDCN patient. Furthermore, we expanded our dataset by integrating publicly available scRNA-seq data of BM samples from 5 BPDCN patients, 5 non-pDC-AML patients, and 5 healthy donors (HD).
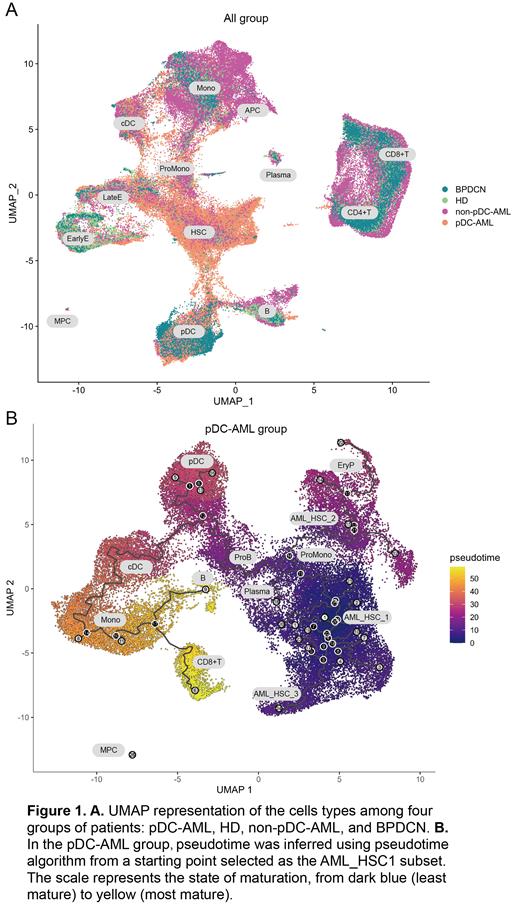
Result: The median pDCs proportion of pDC-AML/MDS was 8.0% (range 2.5-52.0%). As of June 2023, The median overall survival (OS) of patients with pDC-MDS/AML was 10.5 months shorter than non-pDC-AML (21.5 months; P<0.05). In addition, the prognosis of pDC-AML/MDS patients can only be significantly improved after receiving allogenic hematopoietic stem cell transplantation, showing a similar OS to that of non-pDC-AML patients. To further clarified the role of pDCs in these patients, we performed scRNA-seq and acquired 126,693 cells across all samples (8 pDC-AML, 6 BPDCN, 5 non-pDC-AML and 5 HD), and identified 13 clusters based on their transcriptional profile in the overall UMAP (Figure 1 A). Firstly, our single-cell sequencing data revealed significant differences in the proportions of bone marrow pDCs across pDC-AML, HDs, non-pDC-AML, and BPDCN patients. Notably, pDC proportions were significantly elevated in pDC-AML and BPDCN patients. Further differential expression analysis revealed striking differences in the transcriptomes of pDCs across these four groups. When comparing pDC-AML to HDs, we identified 801 upregulated and 150 downregulated genes. Upon comparing pDC-AML to BPDCN, we found 1110 upregulated and 765 downregulated genes, and when comparing pDC-AML to non-pDC-AML, we observed 1674 upregulated and 945 downregulated genes. To explore the functional implications of these differentially expressed genes (DEGs), these findings were explored further through Gene Set Enrichment Analysis (GSEA), and indicated a heightened response to interferon signaling within the pDC cells of pDC-AML patients. Concurrently, we also discovered an increased expression of gene sets associated with tumorigenic pathways (such as KRAS, PTEN, p53, RAPA, etc.) and those related to embryonic stem cell states in pDCs of pDC-AML.
Drawing on our findings and existing research, we hypothesize that interferon signaling might play a pivotal role in the pathogenesis of pDC-AML. This could be achieved through the enhancement of cell proliferation and self-renewal capabilities, thereby facilitating disease progression. This hypothesis is further supported by our pseudotime analysis. After conducting a more detailed subgroup classification of pDC-AML patients, we found a developmental trajectory linking AML_HSC and pDCs specific to pDC-AML patients. Notably, such a correlation was not observed in the non-pDC-AML patient group.
Conclusions: Our study revealed heterogeneity and underscores the crucial role of pDCs in pDC-AML, as we found that these patients had worse OS. We observed upregulation of interferon signaling and tumorigenic pathways in these pDCs. Moreover, pseudotime analysis suggested a potential developmental relationship between pDCs and AML_HSCs. These findings provide invaluable insights for further investigations into pDCs biological properties in pDC-AML pathology and therapy design.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal